BIOC 2351- BIOCHEMISTRY I
NAME: - IBRAHIM LUNAT
I.D. #: - 20053845
SUBJECT: - ESSAY- CRITIQUE ON THE ARTICLE- PHOSPHORYLATION IN YEAST
PHOSPHORYLATION IN YEAST
Background: -
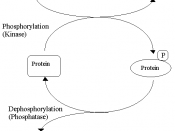
Phosphorylation is the addition of a phosphate (PO4) group to a protein or other organic molecule. Protein phosphorylation in particular plays a significant role in a wide range of cellular processes. In Biochemistry protein phosphorylation has made a significant contribution in researches throughout different biological fields. If we look at the previous researches, in 1906 Phoebus Levene at Rockefeller institute, identified phosphate in the protein Vitellin and after that in 1933 he detected phosphoserine in Casein, a milk protein. But it wasn't until twenty years before Eugene Kennedy described the first 'enzymatic phosphorylation of proteins'.
Phosphorylation of proteins is an important regulatory mechanism which occurs in both prokaryotes and eukaryotes. Enzymes called kinases, which are used in phosphorylation, are involved in this process.
During the process many enzymes and receptors are switched "on" or "off" by phosphorylation and dephosphorylation. Reversible protein phosphorylation provides a major regulatory mechanism in eukaryotic cells. Due to high variability of amino acid residues producing a relatively limited number of experimentally identified phosphorylation sites, there is no reliable prediction of such sites. Phosphorylation usually occurs on serine, threonine and tyrosine residues in eukaryotic proteins. Moreover, in prokaryotic proteins phosphorylation takes place on basic amino acid residues such as histidine, arginine or lysine.
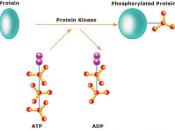
Protein phosphorylation catalysed by protein kinases plays a major role in cellular signaling. There are thousands of distinct phosphorylation sites in a given cell because there are many different kinds of proteins in any cell. It has been estimated that 1/10th to ý of proteins are phosphorylated. Phosphorylation occurs mostly on multiple distinct sites on a given protein. Phosphorylation of any site on a given protein can change function of localization of that protein. Within a protein, phosphorylation can occur on several amino acids. Phosphorylation on serine is most common, followed by threonine.. Protein kinases act on and modify the activity of specific proteins. These are used extensively to transmit signals and control complex processes in cells. Up to 500 different kinases have been identified in humans. A typical eukaryotic cell has the capacity to make about 30,000 different proteins, which catalyze thousands of different reactions involving many metabolites. Cyclin-dependent kinases (CDK) belong to a group of protein kinases originally discovered as being involved in the regulation of the cell cycle.
CDKs phosphorylate proteins on serine and threonine amino acid residues: they are serine/threonine kinases. A cyclin-dependent kinase is activated by association with a cyclin, forming a cyclin-dependent kinase complex.
One of the terms to understand which is used in the article is a 'sequence motif', which is a nucleotide or an amino acid sequence pattern that is widespread and has, or is conjectured to have a biological significance. Within a sequence or database of sequences, researchers search and find motifs using computer-based techniques of sequence analysis, such as BLAST. Bioinformatics field tends to use such techniques. The new technology in market these days is the protein-chip technology which allows thorough analysis of biochemical activities and can be used to analyze protein kinases as well. Protein microarrays making use of new developments in protein engineering and detection have recently emerged. The basic construction of such protein chips has some similarities to DNA chips, such as the use of glass or plastic surface dotted with an array of molecules. With fluorescent markers or other methods of detection revealing the spots that have captured these proteins, protein micro arrays are being used as powerful tools in high-throughput proteomics and drug discovery. At this time, protein-chip technology concentrates on understanding molecular pathways that in turn lead to useful information for drug discovery and which can be applied to molecular diagnostics.
CRITIQUE: -
Realizing the importance of protein phosphorylation which controls many cellular processes, the authors of this article have identified over 4000 phosphorylation events which involved over a thousand proteins. They used the proteome-chip technology to describe the in vitro substrates recognized by the protein kinases. The authors believe that the results will shed light on mechanisms and roles of protein phosphorylation in eukaryotes. This is understood as protein phosphorylation controls many processes in the cells. Moreover yeasts possess many protein kinases and serve as good organisms for study.
The authors analyzed different protein kinases along with A ones which are a family of enzymes whose activity is dependent on the level of cyclic AMP. Protein kinase A has several functions in cell like glycogen and lipid metabolism. A brief note about the protein kinases would have been useful. They then carried out proteome micro arrays on the kinases by using 33-P labeled isotope on ATP. The authors can be commended no their use of negative and positive controls of the experiment. They made the experiment easier by using a large number of proteins and sources for identification of phosphorylation signals. They also used incubated slides in absence of kinases so as to see if any autophosphorylated. This served as a blank. They measured the extent of phosphorylation by algorithmic methods. Although this method may be 70 to 80 percent (according to various sources) efficient, it is relevant here as many kinases need to be analyzed and the method is widely approved in this era of technology. The phosphorylated proteins were marked as positive substrates in the results.
According to the results, since many phosphorylation events occurred, the authors said that each kinase had a certain amount (between 1 and 256) of substrates with an average of forty seven substrates per enzyme. Even though criticizing the authors for not giving accurate results would be incorrect, mention still can be made that an average as such cannot be taken which has a standard deviation so high but for the information of reader it can be accepted. What this means is that an average for kinase activity would not be relevant in this case. The authors mentioned that more than one kinases had about three substrates indicating specificity. There may be an element of doubt in this matter. Enzymes are highly specific molecules. There may have been the same kinase they analyzed but due to improper labeling, gave different results. It is still possible that the enzymes have more than one substrate. The term 'transcription factors were the largest proteins phosphorylated' was not fully understood. They found that the kinases phosphorylated the proteins which resided in the same cell compartment or which came under the same function and this was proven with an example.
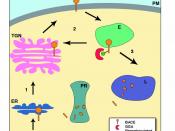
The authors then carried out a test in which they assayed substrates of six kinases to test various other factors. This complex test gave some good results as would be expected. This proved that the kinases controlled protein levels directly or indirectly. The went further to say that using the proteome micro array technology helped them identify many genuine substrates and proved that phosphorylation affected protein levels with a marked difference. Next, they examined the kinases which were closely related to a substrate. They used yeast protein kinase A homologues. They went on to determine if these homologues were functionally redundant. From results they concluded that each homologue verified specificity. More than 3/4th of bovine cAMP-dependent protein kinase targets were also substrates of the homologues. The slides with 90% substrate overlap depicted that each homologue had distinct substrate specificity. A good diagram gave information on how the proteome array test was carried out by in vivo validation. Figure 2(a) showed that TPK1 had highest substrate recognition profile. Figure 2(b) showed how cyclin affected substrate recognition. It was also found that position and structure of the substrate restricted the kinase from phosphorylation.
Later, the 4,200 proteins-substrate phosphorylations were assembled into an in vitro network to be analyzed and a map was created. From this they found that identification of substrate assisted in defining the role of kinase in yeast signaling networks. They said the low overlap between the data sets was due to kinase-substrate interaction had lo binding affinities and were not detectable in the assay. Going in depth with the phosphorylation network, in which the kinase and substrate were present in the same compartment or functional category, resulted in 33% and 18.4% interactions, respectively. They outlined a limitation to this approach of enrichment saying that this approach may give false negatives if the data interpreted were inaccurate or incomplete. The figure of in vivo phosphorylation map given was similar to that of the yeast, Saccharomyces cerevisiae. Then after, to explain how phosphorylation is incorporated in global networks, they combined phosphorylation data with transcription factor binding to generate an integrated regulatory network for yeast. Transcription factor is a protein that binds to specific sequence of DNA and hence controls the transfer of genetic information from DNA to RNA. The number of transcription factors which can be found in an organism increases with the genome size and the larger genomes tend to have more transcription factors per gene. Then , within the network they looked for common regulatory modules. Eight were observed from which six were of high importance statistically. The authors think their study is the first global investigation of protein phosphorylation by protein kinases using an unbiased approach. In the article, the term 'scaffold' refers to ligand-binding domains of proteins, which are engineered into multiple variants capable of binding diverse target molecules with antibody-like properties of specificity and affinity. The authors have also studied previous in vivo phosphorylations, so that would give them an extra edge on the topic they pursued. When combining their data with others, it matched their studies as well. Even though the authors praise proteome chip technology, it has its side effects but on large scale protein purification, it has been proven effective.
Limitation of proteome chips:
An important obstacle in fabricating proteome chips is the need to produce and purify a large number of proteins in a timely manner. In recent years there has been rapid progress in improving the throughput process and implementing alternative approaches. In vitro transcription-translation systems offer an alternative approach for producing proteins. Scientists have taken this process one step further by directly applying in vitro transcription-translation systems to protein chip fabrication. The obvious advantage is that the expensive and time-consuming steps of ORF cloning, protein expression and purification can be eliminated. However, because of inherent problems with the in vitro system, the size of proteins that can be produced is limited, and the reproducibility of the system is not high. One possible limitation of this approach is that problems with protein folding and the integrity of the proteins printed on the chips might sometimes produce false-positives or missed points. However, the possibility of using a range of different DNA probes, each with a specific modification, to profile cellular proteins on the whole-proteome level is a good prospect. If, as shown here, one or two proteins can be identified with each probe that is applied, a substantial database can be built for further biological validation and reference. In the article, the term 'scaffold' refers to ligand-binding domains of proteins, which are engineered into multiple variants capable of binding diverse target molecules with antibody-like properties of specificity and affinity. The authors have also studied previous in vivo phosphorylations so that would give them an extra edge on the topic they pursued. When combining their data with others, it matched their studies as well.
Conclusion: -
Overall, the authors did an excellent job in the analysis of phosphorylation of yeast. They used the latest technology available to create algorithms and networks which helped in analyzing the substrate-kinase relationships. The proteome chips allows rapid and automated solution to protein phosphorylation, saves time and is efficient than other out-dated techniques. The article also gives an insight into what could be further done to take this topic to another level for research, and hence can lay a foundation to it.
References: -
Global analysis of protein phosphorylation in yeast: Jasonâ¦Snyder, 2005 nature publishing group.
Nelson and Cox, Lehninger principles of biochemistry, 5th edition:2008
Koch C, et al. Cell cycle regulated transcription in yeast. 1994 Jun;6(3):451-9. Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. 1993 Apr; 5(2):166-79.
http://www.devicelink.com/ivdt/archive/02/07/002.html
http://www.hosted-webs.com/phosphorylation/
http://en.wikipedia.org/wiki/Phosphorylation
http://www.jbc.org/cgi/content/abstract/252/9/3082
http://biowww.net/detail-1142.html
http://en.wikipedia.org/wiki/AMP-activated_protein_kinase
http://www.rockefeller.edu/labheads/crossf/cdk.html
http://networks.gersteinlab.org/
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2576280
http://www.functionalgenomics.org.uk/sections/resources/protein_arrays.htm
http://genomebiology.com/2001/2/10/reports/0035