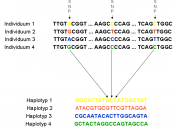
Single Nucleotide PolymorphismDNA sequence variations that occur when a single nucleotide (A, T, C, or G) in the genome sequence is altered. Each individual has many single nucleotide polymorphisms that together create a unique DNA pattern for that person. SNPs promise to significantly advance our ability to understand and treat human disease.
Within a population, SNPs can be assigned a minor allele frequency - the ratio of chromosomes in the population carrying the less common variant to those with the more common variant. Usually one will want to refer to SNPs with a minor allele frequency of âÂÂ¥ 1% (or 0.5% etc.), rather than to "all SNPs" (a set so large as to be unwieldy). It is important to note that there are variations between human populations, so a SNP that is common enough for inclusion in one geographical or ethnic group may be much rarer in another.
SNPs may fall within coding sequences of genes, noncoding regions of genes, or in the intergenic regions between genes.
SNPs within a coding sequence will not necessarily change the amino acid sequence of the protein that is produced, due to redundancy in the genetic code. A SNP in which both forms lead to the same protein sequence is termed synonymous - if different proteins are produced they are non-synonymous. SNPs that are not in protein coding regions may still have consequences for gene splicing, transcription factor binding, or the sequence of non-coding RNA.
SNPs make up 90% of all human genetic variations, and SNPs with a minor allele frequency of âÂÂ¥ 1% occur every 100 to 300 bases along the human genome, on average, where two of every three SNPs substitute cytosine with thymine.
Variations in the DNA sequences of humans can affect how humans develop diseases, respond to pathogens, chemicals, drugs, etc. As a consequence SNPs are of great value to biomedical research and in developing pharmacy products. Because SNPs are inherited and do not change much from generation to generation, following them during population studies is straightforward. They are also used in some forms of genealogical DNA testing.
Detection of SNPA convenient method for detecting SNPs is restriction fragment length polymorphism (SNP-RFLP). If one allele contains a recognition site for a restriction enzyme while the other does not, digestion of the two alleles will give rise to fragments of different length. Currently, the study of existing SNPs is most easily studied using microarrays. Microarrays allow the simultaneous testing of over a thousand separate SNPs and are quickly screened by computer.
Uses of SNPHelps in identifying disease genesSNPs will catapult into the era of personalized medicine, when pharmacogenetics will enable physicians to prescribe drugs based on detailed knowledge of our genotypes.
SNPs are used as genetic marker-the equivalent of landmarks in the human genome. They help in keeping record of the "recombination segments" -blocks of 3000-30,000 base pairs in which SNPs tend to be associated with one another. These blocks are mixed and matched by the process of recombination.
These markers provide:1.Information about a patient's risk for disease2.Insight into the disease process3.Protein targets for novel drug therapiesBenefits of Using SNPs1.A person's SNP pattern is highly unlikely to change over time or as a result of disease.
2.SNP data can be collected from any tissue in the body (not just from diseased tissue).This allows a larger number of samples to be obtained (especially controls) since faster and less invasive procedures are used.
Challenges of Using SNPs1.There are now over one million SNPs known but measuring them all is typically cost-prohibitive. SNP data contain measurements for only a small fraction of known SNPs (typically a few thousand). If prior knowledge is available, focus the SNPs collected to particular region(s) of the genome. Otherwise, choose SNPs to give good overall coverage of the genome.
2.SNP data commonly contain missing values. This can adversely affect many algorithms used for classification tasks. When choosing an algorithm to use, this must be taken into consideration in order to choose an appropriate one.
3.Proper and accurate mining of the SNP data requires instrumentations with super computing facility. Hence, the cost factor takes center stage.
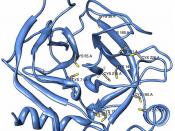
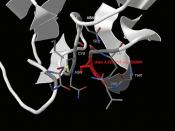
ÃÂ (1,4) galactosyltransferaseÃÂ (1,4) galactosyltransferase (b4GalT-I) is a constitutively expressed, trans-Golgi resident, type II membrane-bound glycoprotein that catalyzes the transfer of galactose to N-acetylglucosamine residues, forming the b4-N-acetyllactosamine (Galb4-GlcNAc) or poly-b4-N-acetyllactosamine structures found in glycoconjugates (15, 16).
ÃÂ4-Galactosyltransferase enzymatic activity is widely distributed in the vertebrate kingdom, in both mammals and nonmammals, including avians (17) and amphibians (unpublished observations) (15).
ÃÂ4GalT-I functions in lactose biosynthesis. In mammals ò4GalT-I has been recruited for a second biosynthetic function, the tissue-specific production of lactose which takes place only in the lactating mammary gland. The synthesis of lactose is carried out by the protein heterodimer assembled from b4GalT-I and the mammalian protein a-lactalbumin (15). The notion that the ò4GalT-I gene has been recruited from the nonmammalian vertebrate pool of constitutively expressed genes for lactose biosynthesis is supported by the observation that the ò4GalT-I ortholog from chicken (15) can also functionally interact with a-lactalbumin in vitro.
The presence of five additional ò4GalT-I related sequence groups (genes) in the human genome, or a total of six genes when ò4GalT-I is included. The family members are designated as ò4GalT-I, -II, -III, -IV, -V and -VI, where ò4GalT-I represents the previously well-characterized ò4GalT recognized to function in lactose biosynthesis (15).
The following diagram indicates the chromosome number and location of each of the gene family members.
FIG 4: Schematic representation of the human 4GalT family members. The transcript representing the gene located on human chromosome 9p13 (4GalT-I) is shown at the top. The five additional family members (4GalT-II through -VI) are shown with their chromosomal location and mRNA size (from Northern blot analysis) noted. The open box indicates coding sequence; the first three numbers indicate the number of amino acids in the stem region, catalytic domain and full-length coding region, respectively. The total number of nucleotides in the coding region is also shown. Since the full-length 5'-untranslated region of each homolog has not been determined, this region is depicted by a dashed line with the number of nucleotides obtained from the most 5'-clone indicated. The thin line at the right indicates the 3'-untranslated region with the number of nucleotides, available from the EST clones shown. As three of the homologs (4GalT-II,-V, and -VI) do not contain a consensus polyadenylation signal sequence (An), the predicted length of the 3'-untranslated region is given in italics. The sequence of 4GalT-II and -VI that was obtained by RACE, is 5'of the solid arrowhead. Superimposed on each mRNA is the position of the transmembrane domain (solid box) and the position of each Cys residue. The position, in 4GalT-I of the only intramolecular disulfide bond, Cys130 and Cys243 is indicated. Cys338 in the 4GalT-I sequence is replaced by a Tyr in each family member.
Identification of large family of ÃÂ (1,4) galactosyltransferaseSeveral groups independently used the emerging EST database information in 1997 to identify a group of human cDNA sequences with similarities to the classical ÃÂ4Gal-T (designated ÃÂ4Gal-T1) (18, 19). Within 1 year, five novel human ÃÂ4Gal- T genes designated ÃÂ4Gal-T2 to -T6 were identified cloned, and enzymatic functions of their recombinant proteins demonstrated (18, 19).The two genes, ÃÂ4Gal-T5 and -T6, were identified by traditional cloning strategies as well as computer cloning (18). Recently, a seventh homologous gene designated ÃÂ4Gal-T7 was identified by the computer cloning strategy (18, 20). Its homology has not been established yet.
BibliographyReferences1.Serum galactosyltransferase isoform changes in rheumatoid arthritis, Alavi et al., J Rheumatol. 2004 Aug; 31(8):1513-20.
2.New Insights into rheumatoid arthritis associated glycosylation changes, Alavi et al., Adv Exp Med Biol. 2005; 564:129-38.
3.Functional interaction between the SSeCKS scaffolding protein and the cytoplasmic domain of ÃÂ1,4-galactosyltransferase, Wassler et al., Journal of Cell Science 114, 2291-2300 (2001)4.Tumor Necrosis Factor-ñ Microsatellite Polymorphism Association with Rheumatoid Arthritis in Indian patients, Agrawal, et al, Archives of Medical Research 36 (2005) 555-559.
5.Changes in Normal Glycosylation Mechanisms in Autoimmune Rheumatic Disease, Axford, et al., Glycosylation Mechanisms and Auloimmune Rheumatic Disease.
6.Structural analysis of the N-glycans from human immunoglobulin Al: comparison of normal human serum immunoglobulin Al with that isolated from patients with rheumatoid arthritis, Field, et al, Biochem. J. (1994) 299, 261-275.
7.B lymphocyte galactosyltransferase protein in normal individuals and in patients with rheumatoid arthritis, Keusch, et al, Glycoconjugate Journal 15, 1093-1097 (1998)8.dbSNP: The NCBI database of genetic variation, Sherry, et al, Glycoconjugate Journal 15, 1093-1097 (1998)9.A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms, The International SNP Map Working Group, (2001) Nature, 409: 928-93310.High-Throughput Identification, Database storage and Analysis of SNPs in EST Sequences, Useche et al. (2001), Genome Informatics 12: 194-20311.A general approach to single-nucleotide polymorphism discovery, Marth et al. (1999), Nature Genetics, 452-45612.dbSNP-Database for Single Nucleotide Polymorphisms and other Classes of Minor Genetic Variation. Sherry, S.T., Ward, M. and Sirotkin, K. (1999), Genome Research, 9, 677-67913.Reading Bits of Genetic Information: Methods for Single-Nucleotide Polymorphism Analysis, Landegren et al. (1998), Genome Research, 8:769-77614.Variations on a theme: cataloging human DNA sequence variation. Collins, F.S., Guyer, M.S. & Chakravarti, A., (1997), Science 278, 1580-158115.The expanding b4-galactosyltransferase gene family: messages from the databanks.
Neng-Wen Lo, Joel H.Shaper, Jonathan Pevsner and Nancy L.Shaper, (1998), MD 21287-8937, USA.
16.Glycosylation pathway in the biosynthesis of nonreducing terminal sequences oligosaccharides of glycoproteins, Beyer,T.A. and Hill,R.L., (1968), Horowitz,M. (ed.), The Glycoconjugates. Vol. III, Academic Press, New York, pp. 25-45.
17.The chicken genome contains two functional nonallelic b1,4-galactosyltransferase genes: chromosomal assignment to syntenic regions tracks fate of the two gene lineages in the human genome, Shaper,N.L., Meurer,J.A., Joziasse,J.H., Chou,T.-D.D., Smith,E.J., Schnaar,R.L and Shaper,J.H., (1997), J.Biol. Chem., 272, 31389-31399.
18.Identifcation and characterization of large galactosyltransferase genefamilies: galactosyltransferases for all functions, Margarida Amado, Raquel Almeida, Tilo Schwientek, Henrik Clausen, (1999), Biochimica et Biophysica Acta 1473 (1999) 35-53.
19.A Family of human ÃÂ4-galactosyltransferases: cloning and expression of two novel UDP-galactose: ÃÂ-n-acetylglucosamine ÃÂ1,4-galactosyltransferases, ÃÂ4Gal-T2and ÃÂ4Gal-T3, R Almeida, M. Amado, L. David, S.B. Levery, E.H. Holmes, G. Merkx, A.G. van Kessel, H. Hassan, E.P. Bennett, H. Clausen, J. Biol. Chem. 272 (1997) 31979-31992.
20.Cloning and expression of a proteoglycan UDP-galactose:ÃÂ-xylose ÃÂ1,4-galactosyltransferaseI. A seventh member of the human ÃÂ4-galactosyltransferase gene family, R. Almeida, S.B. Levery, U. Mandel, H. Kresse, T. Schwientek, E.P. Bennett, H. Clausen, J. Biol. Chem. 274 (1999) 26165-26171.
21.Use of site-directed mutagenesis to identify the galactosyltransferase binding sites for UDP-galactose, H. Zu, M.N. Fukuda, S.S. Wong, Y. Wang, Z. Liu, Q.Tang, H.E. Appert, Biochem. Biophys. Res. Commun. 206 (1995) 362-369.Mizuochi T., Taniguchi T, Shimizu A. and Kobata A. (1982), J. Immunol. 129, 2016-2020.
22.Malhotra R, Wormald M.R., Rudd P., Fischer P.B., Dwek R.A. and Sim R.B. (1995), Nature Med. 1.237-243.
23.Roitt IM, Dwek R.A., Parekh R.B., Rademacher T.W., Alavi A, Axford J.S., Bodman K., Bond A., Cooke A., Hay F.C., et.al (1988). Recenti Progressi medicina 79. 314-317.
24.Abnormalities in IgG glycosylation and immunological disorders, Alavi A., Axford J.S., (1996) pp. 149-169, John Wiley and sons ltd, London.
25.Podolsky D.K., Weiser M.W., Westwood J.C. and Gammon M., (1997), J. Biol. Chem. 252. 1807-1813.
26.Serum galactosyl transferase as a marker of disease activity in rheumatoid arthritis, Azita Alavi, Axford J.S., (1997), Biochemical society transactions 25., 313.
27.Role of PTPN22 in type 1 diabetes and other autoimmune diseases, Bottini N, Vang T, Cucca F, Mustelin T., (2006 May 10), Semin Immunol.